Using CRISPR for Microbial Genome Editing
As in mammalian cell lines, CRISPR/Cas9 technology can be used to generate knock-in and knock-out mutations in microbes. While HR frequency is generally lower in microbes than mammalian cells, alternative techniques can be utilized to improve recombination efficiency. Optimized gene editing in microbes can be achieved by coupling CRISPR/Cas9 technology with λ Red recombination techniques. The λ Red system utilizes bacteriophage recombinases to mediate recombination of homologous sequences as short as 30 base pairs.
In this case study, λ Red – CRISPR/Cas9 was used to knock-out cadA in the BL21 E.coli strain. The CadA protein is a component of lysine decarboxylase, an enzyme that helps bacteria survive in acidic environments. After editing, Sanger sequencing and colony PCR screening was used to confirm that the cadA gene was successfully knocked-out (Figure 1).
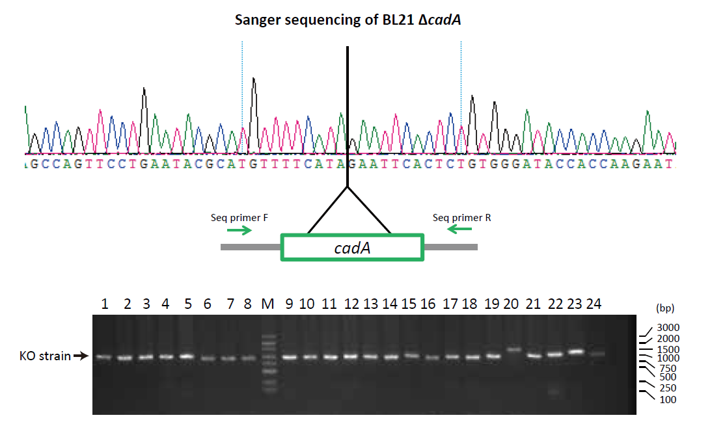
Figure 1: Knock-out of cadA verified by Sanger sequencing and colony PCR
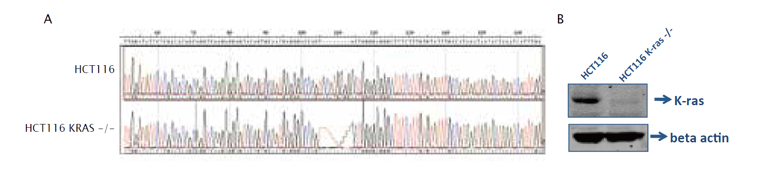
Figure 2: KRAS loss-of-function determined by Sanger sequencing and Western blotting
- Like (1)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2025 MolecularCloud




