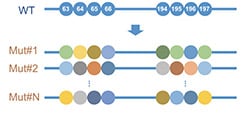
Top 3 Advantages of Precision Mutant Libraries for Protein Engineering
What are Precision Mutant Libraries?
Imagine finding a happy medium between random and rational protein engineering; this sums up “Precision Mutant Libraries.” By taking advantage of Next-Generation semiconductor-based DNA Synthesis Technology operating within an automated platform, you can introduce exact nucleotide changes, allowing testing your predictions and unraveling connections between your protein’s sequence, structure, and function.
Vesna Mitchell, Ph.D.,
Codexis Protein Engineering Experts
"We really like to know what is in our libraries; we also like to reduce our screening efforts and get the maximum amount of information. If you have random changes in your library, you can't quite learn as much from them." "If we can screen less and learn more, that's always a good thing.
Reduce your screening burden - If you have embarked on protein engineering projects before, then you know the hardships of seemingly endless screening efforts.
Random mutagenesis approaches have led to amazing discoveries. Yet, for all their strengths, the main caveat remains…. Huge mutant libraries requiring extensive, well-designed, and carefully executed selection methods. An almost herculean task!
Possu Huang, Ph.D.,
Assistant Professor, Stanford University
"if we form a hypothesis and we build computational models of these proteins, the fact that you can cover exactly the distribution, positions, and mutation sites, it can actually feedback into our hypothesis much better than dealing with degenerate codons."
Choose your codons - Ever wished you could make only desired changes in unique protein domains to more quickly and directly test your hypothesis?
Whether you want to change a specific codon, contiguous codons, or multiple codons in different protein regions, precise control over oligonucleotide synthesis allows you to test only desired mutations. Absolute control over oligonucleotide synthesis has the added benefit of eliminating stop codons that would otherwise result in fruitless pursuits during screening.
Meredith Jackrel, Ph.D.,
Assistant Professor, Washington University
"they achieved quite high diversity at every position and we have nearly all of the 4,940 variants represented" "Even if we would have done this using degenerate oligonucleotides we wouldn't have been able to achieve this even representation due to inherent codon bias of the genetic code."
Avoid bias - One of the biggest challenges when creating libraries with degenerate codons (e.g., NNK or NNS; where N is A, C, G, or T, K is G or T, and S is C or G) is redundancy, which results in mutant libraries with skewed amino acid distribution.
You may blame the redundancy of the genetic code, where amino acid representation is unequal. Ultimately, while degenerate oligos diminish redundancy, resulting libraries are still biased, and without careful screening, unique variants may be missed.
At Your Service
Are you ready to take these unparalleled advantages offered by our Precision Mutant Libraries (PMLs)? Visit us at Explore PML Service!
Precision Mutant Library Options
Site-Saturation Mutagenesis
Saturation Scanning Mutagenesis
Combinatorial Mutagenesis
- Like (3)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2025 MolecularCloud