Sanger Sequencing vs. Next-Generation Sequencing (NGS)
Evolution of DNA sequencing
DNA sequencing is the process of identifying the exact order of nucleotide bases (i.e., Adenine, Cytosine, Guanine, and Thymine) encoding specific genomic information. DNA sequencing technology rapidly advanced from its inception in the 1970s with the work of Frederick Sanger, who sequenced the first full genome based on the “plus and minus method” (aSanger et al. 1977). Ultimately, the Sanger chain termination or dideoxy method, also first reported in 1977, provided the foundation for fast-paced growth in DNA sequencing technologies and enabled sequencing the human genome for the first time in 2001 (bSanger et al. 1977).
Next-Generation sequencing (NGS) technologies evolved from the need to sequence larger volumes of genetic material faster and at a lower cost. Over the years, different NGS methods and platforms have been developed based on unique chemistries and detection methods (e.g., Pyrosequencing, Reversible-dye terminator, and Proton detection) (Garrido-Cardenas et al. 2017). Nevertheless, NGS or second-generation technologies share their reliance on massively parallel processing and the potential to sequence thousands to millions of DNA strands simultaneously.
Next-generation technologies have undoubtedly reduced DNA sequencing costs and expedited high-quality genome sequencing. Nevertheless, Sanger sequencing remains a method of choice, especially for the analysis of low-volume DNA sequences, as it provides high-quality DNA sequencing data for regions of up to 1,000 bases.
How does Sanger sequencing work?
Sanger sequencing, or the enzymatic chain termination method, is based on the use of fluorophore-labeled dideoxynucleotides (ddNTPs) in combination with regular deoxynucleotides (dNTPs). Because ddNTPs lack a hydroxyl group needed for nucleotide binding, the addition of ddNTPS by the DNA polymerase during chain extension terminates the DNA strand elongation. Reiteration of primer annealing and DNA extension cycles results in fragments with a fluorophore-labeled nucleotide at each position, thereby identifying every nucleotide in the DNA template (Crossley et al. 2020).
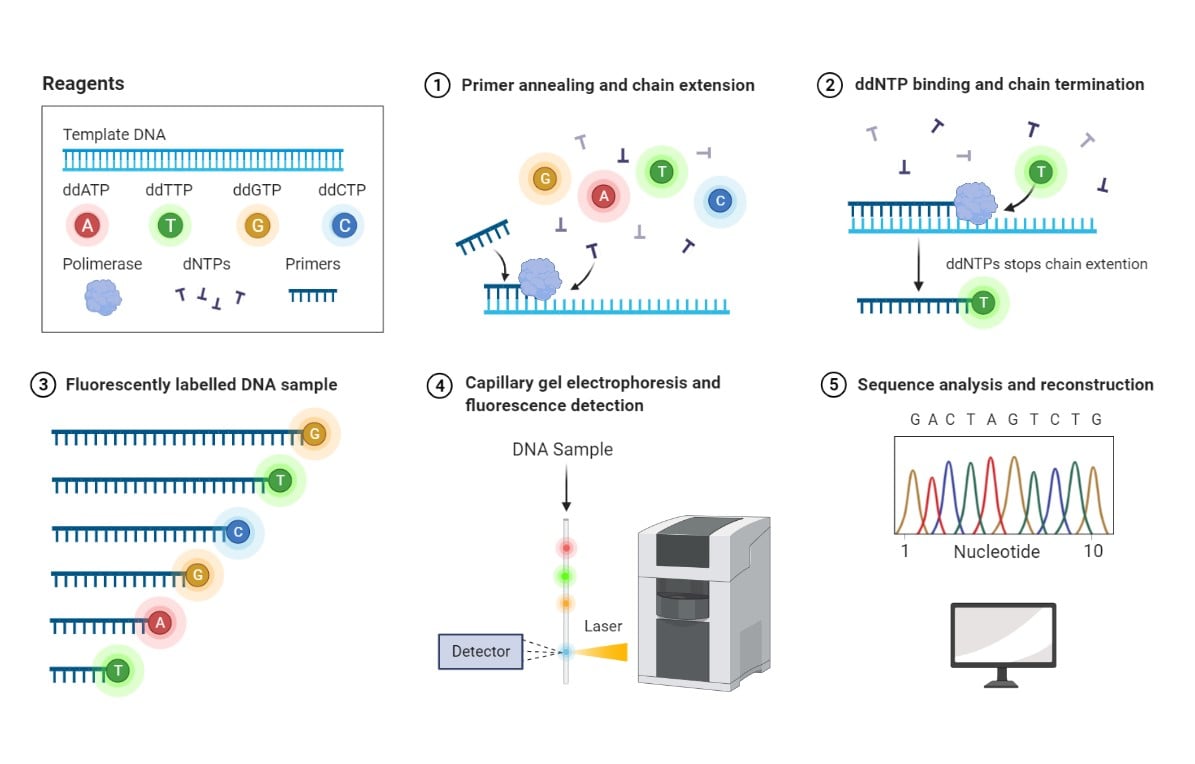
Sanger sequencing method. 1- Deoxynucleotides (dNTPs) and a low concentration of fluorophore-labeled dideoxynucleotides (ddNTPs) are supplied together with DNA template and primers. Polymerase catalyzes the DNA chain extension by random incorporation of available nucleotides. 2- Binding of a fluorophore-labeled ddNTP leads to termination of chain extension. 3- DNA fragments of various lengths and labeled with different fluorophores are generated. 4- Labeled DNA fragments are separated based on molecular mass by capillary gel electrophoresis, and their associated fluorescence is detected by a Charge-Coupled Device (CCD). 5- A chromatogram is generated based on peak fluorescence detected, corresponding to fluorophore-labeled ddNTPs incorporated at each position of the DNA sequence. Reprinted from “Sanger Sequencing”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
You could check the full content here: https://www.genscript.com/gene-news/sanger-sequencing-vs-next-generation-sequencing.html
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2025 MolecularCloud




DNA