CRISPR/Cas13: Evolving Biomedical Applications
The CRISPR/Cas13 family consists of four nuclease subtypes, Cas 13a-d. Unlike CRISPR/Cas9 and CRISPR/Cas12, which target DNA, CRISPR/Cas13 is guided by RNA to target specific RNA sequences for degradation. In addition to targeted RNA degradation, Cas13 leads to “collateral RNA cleavage,” an activity demonstrated in vitro and in bacteria but not in mammalian cells (Abudayyeh et al. 2016 and 2017).
Harnessing the versatility of Cas13 unique activities, recently developed strategies target these nucleases to viral RNA in support of diagnostics and treatments. These strategies have the potential to broadly impact the way we currently diagnose and treat viral infections.
CRISPR/Cas13 in Viral Detection
During the COVID-19 pandemic, the unique functions of Cas13a were leveraged for the development of a SARS-CoV-2 viral detection assay. The SHERLOCK assay provides a specific, sensitive and rapid test for the detection of SARS-CoV-2 RNA. In this assay, viral RNA is pre-amplified to support sensitive detection by the Cas13a/crRNA complex. Detection of viral RNA by Cas13a unleashes its RNAse or collateral cleavage activity upon an RNA reporter generating a fluorescent signal (Gootenberg et al. 2017, Kellner et al. 2019).
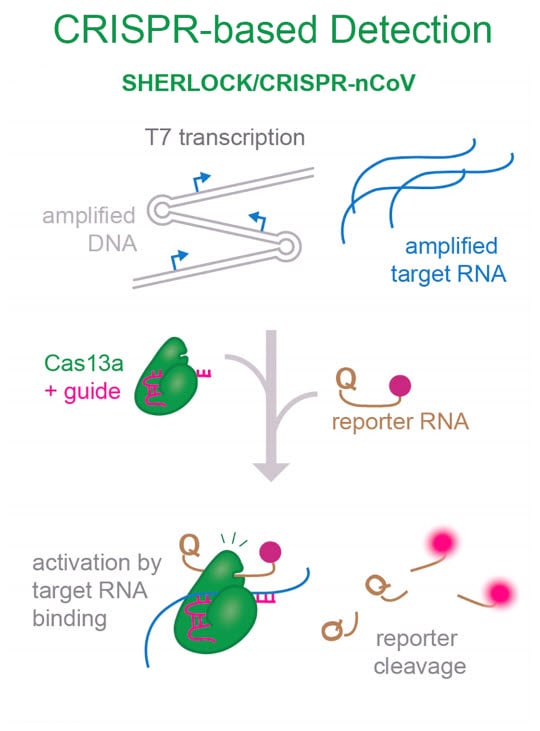
SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unlocking) is a sensitive CRISPR-based viral detection assay that supports rapid detection of SARS-CoV-2 (<1 hour). In this assay, target RNA is pre-amplified to ensure high sensitivity for viral RNA detection. Adapted from Esbin et al., 2020. Figure 8 was modified by removing the sections on DETECTR/AIOD-CRISPR/CDetection and lateral dipstick readout. https://creativecommons.org/licenses/by/4.0/
More recently, a new diagnostic method has been developed termed SATORI (CRISPR-based amplification-free digital RNA detection). This new assay combines the use of CRISPR/Cas13a and a microchamber-array device to sensitively detect SARS-CoV-2 RNA in the absence of a pre-amplification step (Shinoda et al. 2021). Similar to the SHERLOCK test, the readout is based on the cleavage of a fluorescent RNA reporter. Shinoda and colleagues demonstrated that this approach enabled fast detection (< 5 min) of target SARS-CoV-2 N-gene RNA with a sensitivity that varied according to the specific crRNA used (LOD values ~9-100 fM). Combined use of crRNAs with different SARS-CoV-2 N-gene RNA target specificities substantially improved sensitivity (LOD values ~5 fM). Importantly, Shinoda et al. demonstrated the applicability of SATORI to the analysis of raw clinical samples.
Overall, the most significant advantage of this approach is that it bypasses potential amplification-related errors that may confound diagnosis. Nevertheless, compared to SHERLOCK’s sensitivity for SARS-CoV-2 detection, SATORI is ~1000 times less sensitive (Kellner et al. 2019).
CRISPR/Cas13 in Viral Treatment
The ability of CRISPR/Cas13a and Cas13d to target viral RNA in vitro has been demonstrated previously (Freije et al. 2019, Abbott et al. 2020). These studies established the effectiveness of a CRISPR/Cas13 based approach in reducing viral RNA for several viruses, including influenza A and SARS-CoV-2.
In a new approach, the RNA targeting and RNAse properties of CRISPR/Cas13a have been applied in vivo to control respiratory RNA viruses (Blanchard et al. 2021). For this application, delivery of the Cas13a nuclease relied on synthetic mRNA encoding Leptotrichia buccalis, LbuCas13a. Blanchard and colleagues reasoned that the use of Cas13a mRNA would support transient Cas13 protein expression and reduce the potential for adverse immunological reactions.
Following in vitro evaluation of crRNAs targeting specific influenza A and SARS-CoV-2 viral RNA sequences, Cas13a mRNA and crRNAs were delivered as part of a polymer complex or polyplex through inhalation into the lungs of infected rodents (Patel et al. 2019). Blanchard and colleagues were successful at demonstrating that CRISPR/Cas13a reduced viral RNA in vivo, in mice and hamsters infected with influenza A and SARS-CoV-2 virus, respectively.
Overall, the huge advantage provided by CRISPR/Cas13 based strategies in diagnostics and treatment of viral infections is its flexibility and potential to target a broad range of viral sequences and pathogenic viruses. With this goal in mind, a group at Stanford University has developed a bioinformatics tool (http://crispr-pacman.stanford.edu/) to share information on Cas13d crRNAs that effectively target a broad range of RNA viruses (Lin et al. 2021).
Reference
Abbott, T. R. et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell (2020) doi:10.1016/j.cell.2020.04.020.
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science (80-. ). (2016) doi:10.1126/science.aaf5573.
Abudayyeh, O. O. et al. RNA targeting with CRISPR-Cas13. Nature (2017) doi:10.1038/nature24049.
Blanchard, E. L. et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. (2021) doi:10.1038/s41587-021-00822-w.
Esbin, M. N. et al. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA (2020) doi:10.1261/rna.076232.120.
Freije, C. A. et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell (2019) doi:10.1016/j.molcel.2019.09.013.
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science (80-. ). (2017) doi:10.1126/science.aam9321.
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O. & Zhang, F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. (2019) doi:10.1038/s41596-019-0210-2.
Lin, X. et al. A comprehensive analysis and resource to use CRISPR-Cas13 for broad-spectrum targeting of RNA viruses. Cell Reports Med. (2021) doi:10.1016/j.xcrm.2021.100245.
Patel, A. K. et al. Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. (2019) doi:10.1002/adma.201805116.
Shinoda, H., Taguchi, Y., Nakagawa, R. et al. Amplification-free RNA detection with CRISPR–Cas13. Commun Biol 4, 476 (2021). doi.org/10.1038/s42003-021-02001-8.
- Like (4)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2025 MolecularCloud



