COVID-19 Pathogenesis: Uncovering New Molecular Mechanisms
To no one's surprise by now, SARS-CoV-2 is very efficient at infecting our lungs. Once there, it wreaks havoc, attacking airway and alveolar epithelial cells preferentially. But what are the molecular mechanisms underscoring the reported lung damage associated with severe COVID-19? By leveraging phosphoproteomics and genome-wide analysis, recent studies provide insights on different mechanisms and molecular pathways that may serve as therapeutic targets.
Dysregulated Pathways in Alveolar Damage
Within the distal lung, which comprises the terminal and respiratory bronchioles and alveolar ducts, SARS-CoV-2 shows a preference for infecting ACE2 expressing alveolar epithelial type 2 (AT2) cells (Jain and Sznajder 2007, Hou et al. 2020). Investigators have recognized that alveolar and AT2 cell damage drives the acute respiratory distress syndrome (ARDS) frequently occurring in severe COVID-19 cases.
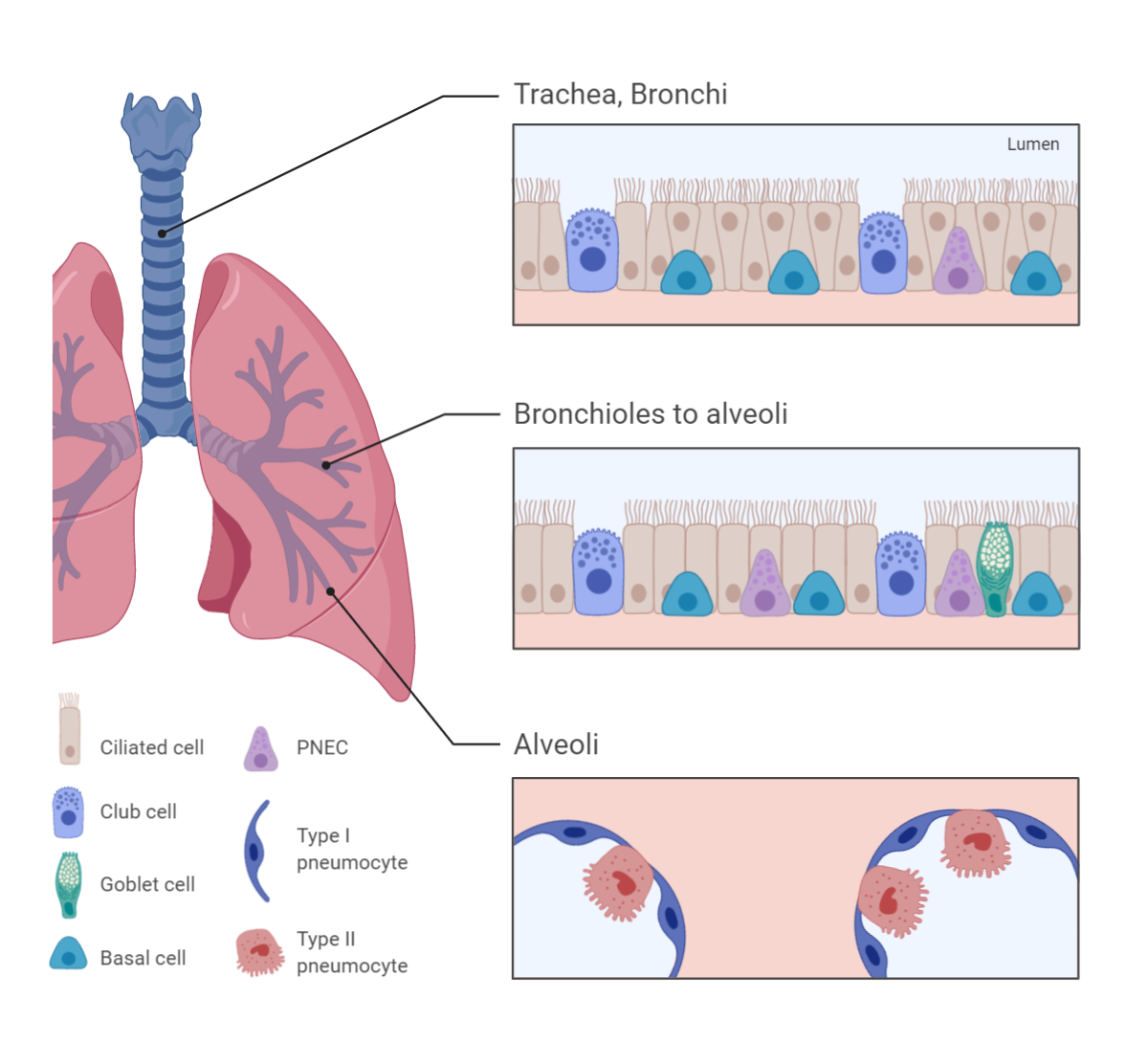
Reprinted from "Respiratory Epithelium" by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
Type II pneumocytes or alveolar type II (AT2) cells are cuboidal cells that occupy less than 10% of the alveolar surface. AT2 cells produce lipoprotein surfactants necessary for proper lung gas exchange function.
To understand the molecular mechanisms leading to AT2 cell injury, investigators used an in vitro model where AT2 cells derived from human induced pluripotent stem cells (iPSCs) were infected with SARS-CoV-2. Proteomics analysis allowed them to uncover diverse molecular pathways involved in COVID-19 lung damage. Hekman et al. identified early responses to SARS-CoV-2 infections, which were shaped by changes in the levels and phosphorylation of host proteins. The identified proteins were involved in immunity, cell proliferation, and survival. Significantly, over 1,000 lung-specific proteins were differentially phosphorylated in response to SARS-CoV-2 infection. Dysregulated pathways included those controlling mRNA splicing, nuclear integrity, metabolism, and apoptosis. Investigators identified several kinases such as CSNK1E, CDK2, EEF2K RPS6KA3, CDK1/2, and MAPK14 susceptible to SARS-CoV-2 and promoting abnormal signaling early during infection. Of relevance to alveolar cells, investigators found abnormal regulation of tight junction integrity. Based on these findings, a Phase 3 clinical trial is underway to evaluate the efficacy of a potent antiviral and MAPK inhibitor, losmapimod, for COVID-19 treatment.
Genomic Determinants of Severe COVID-19 Susceptibility
Severe COVID-19 is distinguishable from mild disease by hypoxemia, lung inflammation, and alveolar damage. Response to anti-inflammatory treatment is also a differentiator between severe and mild COVID-19. For example, only patients with severe disease benefit from corticosteroid treatment. Therefore, to understand the genetic basis for differential responses to SARS-CoV-2 infection, investigators compared patients with severe COVID-19 to healthy controls in a genome-wide association study (GenOMICC study) in the UK. By following this unbiased approach, Pairo-Castineira et al. 2020 identified genetic associations for respiratory failure in severe COVID-19 cases.
First, Pairo-Castineira et al. identified molecular factors involved in the early response to COVID-19 as exemplified by IFN and its downstream target, OAS3. During the early innate immune response, OAS enzymes are involved in viral RNA degradation. This finding points towards a critical role for OAS enzymes' antiviral activity in guarding against severe COVID-19 (Yung Choi et al. 2015). Additionally, factors such as DPP9, TYK2, and CCR2 were linked to the severity of lung inflammatory responses.
Overall, these new studies have proved to be useful in identifying actionable targets for testing the efficacy of existing drugs against COVID-19. Investigators believe that identifying molecular factors involved in the inflammatory phase may provide more effective treatment opportunities to prevent lung damage.
Reference
Choi, U. Y. un., Kang, J. S., Hwang, Y. S. ahn. & Kim, Y. J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Experimental & molecular medicine (2015) doi:10.1038/emm.2014.110.
Hekman, R. M. et al. Actionable Cytopathogenic Host Responses of Human Alveolar Type 2 Cells to SARS-CoV-2. Mol. Cell (2020) doi:10.1016/j.molcel.2020.11.028.
Hou, Y. J. et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell (2020) doi:10.1016/j.cell.2020.05.042.
Jain, M. & Sznajder, J. I. Bench-to-bedside review: Distal airways in acute respiratory distress syndrome. Critical Care (2007) doi:10.1186/cc5159.
Pairo-Castineira et al. Genetic mechanisms of critical illness in Covid-19. Nature (2020) doi:10.1038/s41586-020-03065-y.
- Like (3)
- Reply
-
Share
About Us · User Accounts and Benefits · Privacy Policy · Management Center · FAQs
© 2025 MolecularCloud



